- Компании
- Takeda. О компании, буклеты, каталоги, контакты
- Olympus. О компании, буклеты, каталоги, контакты
- Boston Scientific. О компании, буклеты, каталоги, контакты
- Pentax. О компании, буклеты, каталоги, контакты
- Fujifilm & R-Farm. О компании, буклеты, каталоги, контакты
- Erbe. О компании, буклеты, каталоги, контакты
- Еще каталоги
- Мероприятия
- Информация
- Обучение
- Классификации
- Атлас
- Quiz
- Разделы
- Пациенту
QR-код этой страницы
Для продолжения изучения на мобильном устройстве ПРОСКАНИРУЙТЕ QR-код с помощью спец. программы или фотокамеры мобильного устройства
Статьи: Билиарная микробиота и заболевания желчных путей
| Авторы: | И.Д. Клабуков , А.В. Люндуп, Т.Г. Дюжева 1 2017г. |
| Об авторах: |
1. Первый Московский государственный медицинский университет им. И.М. Сеченова Минздрава России, Институт регенеративной медицины, Москва, Российская Федерация |
Аннотация:
Традиционно считалось, что желчный проток стерилен, а присутствие в желчи микроорганизмов является маркером патологического процесса. Подобное предположение подтверждалось безуспешностью выделения бактериальных штаммов из нормального желчного протока. В настоящей работе приводится обоснование феномена нормальной микробиоты желчевыводящих путей как отдельного слоя, который защищает желчные пути от колонизации экзогенными микроорганизмами. Раскрывается возможное использование метагеномных данных для профилактики инфекционных заболеваний и послеоперационных осложнений при реконструктивных вмешательствах. Методы сохранения гомеостаза экосистемы нормальной билиарной микробиоты могут быть использованы для предотвращения гепатобилиарных заболеваний и лечения воспалительных заболеваний желчевыводящих путей.
Вывод:
В настоящее время имеется достаточно научных данных, обосновывающих актуальность исследования билиарной микробиоты на уровне метагенома (билиарного микробиома).
Существующие методы количественной оценки состава микробиоты с использованием метагеномного секвенирования, последующей биоинформатической обработки и анализа функционала микробиома билиарной системы позволяют выявлять разнообразие микробного
сообщества, населяющего желчные пути в норме, а также характерные микробиомные биомаркеры при наличии патологии. Такие результаты потенциально могут быть использованы для профилактики и терапии микробиотоассоциированных заболеваний желчевыводящей системы.
Все эти данные являются основой для рассмотрения влияния микробиоты на лечение заболеваний не только желчных протоков, но и других органов.
Список литературы:
2. Лоранская И.Д., Ракитская Л.Г., Малахова Е.В., Мамедо-ва Л.Д. Лечение хронических холециститов // Лечащий врач. — 2006. — №6 — С. 12-17. [Loranskaya ID, Rakitskaya LG, Mal-akhova EV, Mamedova LD. Management of chronic cholecystits. Practitioner. 2006;(6):12-17. (In Russ).]
3. Merritt ME, Donaldson JR. Effect of bile salts on the DNA and membrane integrity of enteric bacteria. J Med Microbiol. 2009;58(12):1533—1541. doi: 10.1099/jmm.0.014092-0.
4. Ljungh A, Wadstrom T. The role of microorganisms in biliary tract disease. Curr Gastroenterol Rep. 2002;4(2):167—171. doi: 10.1007/ s11894-002-0055-6.
5. Csendes A, Burdiles P, Maluenda F, et al. Simultaneous bac-teriologic assessment of bile from gallbladder and common bile duct in control subjects and patients with gallstones and common duct stones. Arch Surg. 1996; 131 (4):389—394. doi: 10.1001/arch-surg.1996.01430160047008.
6. Hardy J, Francis KP, DeBoer M, et al. Extracellular replication of Listeria monocytogenes in the murine gall bladder. Science. 2004;303(5659):851—853. doi: 10.1126/science.1092712.
7. Wu T, Zhang Z, Liu B, et al. Gut microbiota dysbiosis and bacterial community assembly associated with cholesterol gallstones in large-scale study. BMC Genomics. 2013;14:669. doi: 10.1186/14712164-14-669.
8. Liang T, Su W, Zhang Q, et al. Roles of sphincter of oddi laxity in bile duct microenvironment in patients with cholangiolithiasis: from the perspective of the microbiome and metabolome. J Am Coll Surg. 2016;222(3):269—280 e210. doi: 10.1016/j.jamcoll-surg.2015.12.009.
9. Jeffery IB, Claesson MJ, O'Toole PW, Shanahan F. Categorization of the gut microbiota: enterotypes or gradients? Nat Rev Microbiol. 2012;10(9):591—592. doi: 10.1038/nrmicro2859.
10. Mowat AM, Agace WW. Regional specialization within the intestinal immune system. Nature Reviews Immunology. 2014;14(10):667— 685.
11. Verdier J, Luedde T, Sellge G. Biliary mucosal barrier and microbiome. Viszeralmedizin. 2015;31(3):156—161. doi: 10.1159/000431071.
12. Hazrah P, Oahn KT, Tewari M, et al. The frequency of live bacteria in gallstones. HPB (Oxford). 2004;6(1):28-32. doi: 10.1080/13651820310025192.
13. Manichanh C, Borruel N, Casellas F, Guarner F. The gut microbiota in IBD. Nat Rev Gastroenterol Hepatol. 2012;9(10):599-608. doi: 10.1038/nrgastro.2012.152.
14. Zhao LP. The gut microbiota and obesity: from correlation to causality. Nature Reviews Microbiology. 2013;11(9):639—647. doi: 10.1038/nrmicro3089.
15. Sekirov I, Russell SL, Antunes LCM, Finlay BB. Gut microbiota in health and disease. Physiol Rev. 2010;90(3):859-904. doi: 10.1152/ physrev.00045.2009.
16. Zeller G, Tap J, Voigt AY, et al. Potential of fecal microbio-ta for early-stage detection of colorectal cancer. Mol Syst Biol. 2014;10(11):766. doi: 10.15252/msb.20145645.
17. Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148(6):1258-1270. doi: 10.1016/j.cell.2012.01.035.
18. Lu LJ, Liu J. Human microbiota and ophthalmic disease. Yale J Biol Med. 2016;89(3):325-330.
19. Josefowicz SZ, Niec RE, Kim HY, et al. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature. 2012;482(7385):395-399. doi: 10.1038/nature10772.
21. Marieb EN, Hoehn K. Human anatomy and physiology. 7th ed. Pearson; 2007. P. 498-499.
22. Balemba OB, Salter MJ, Mawe GM. Innervation of the extrahepatic biliary tract. Anat Rec A Discov Mol CellEvolBiol. 2004;280(1):836-847. doi: 10.1002/ar.a.20089.
23. Hendrickx AP, Top J, Bayjanov JR, et al. Antibiotic-driven dysbiosis mediates intraluminal agglutination and alternative segregation of Enterococcus faecium from the intestinal epithelium. MBio. 2015;6(6):e01346-01315. doi: 10.1128/mBio.01346-15.
24. Li H, Limenitakis JP, Fuhrer T, et al. The outer mucus layer hosts a distinct intestinal microbial niche. Nat Commun. 2015;6:8292. doi: 10.1038/ncomms9292.
25. Savage DC, Dubos R, Schaedler RW. The gastrointestinal epithelium and its autochthonous bacterial flora. J Exp Med. 1968;127(1):67— 76. doi: 10.1084/jem.127.1.67.
26. McFarland LV. Update on the changing epidemiology of Clostridium difficile-associated disease. Nat Clin Pract Gastroenterol Hepatol. 2008;5(1):40—48. doi: 10.1038/ncpgasthep1029.
27. Cohen MB. Clostridium difficile infections: emerging epidemiology and new treatments. J Pediatr Gastroenterol Nutr. 2009;48 Suppl 2:S63—65. doi: 10.1097/MPG.0b013e3181a118c6.
28. Swidsinski A, Loening-Baucke V, Lochs H, Hale LP. Spatial organization of bacterial flora in normal and inflamed intestine: a fluorescence in situ hybridization study in mice. World J Gastroenterol. 2005;11(8):1131—1140. doi: 10.3748/wjg.v11.i8.1131.
29. Limoli DH, Jones CJ, Wozniak DJ. Bacterial extracellular poly-saccharides in biofilm formation and function. Microbiol Spectr. 2015;3(3). doi: 10.1128/microbiolspec.MB-0011-2014.
30. de Vos WM. Microbial biofilms and the human intestinal microbiome. NPJ Biofilms Microbiomes. 2015; 1 (1): 15005. doi: 10.1038/npjbiofilms.2015.5.
31. Chen WG, Liu FL, Ling ZX, et al. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS One. 2012;7(6):e39743. doi: 10.1371/journal. pone.0039743.
32. Swidsinski A, Loening-Baucke V. Functional structure of intestinal microbiota in health and disease. In: Fredricks DN, editor. Human
178 microbiota: how microbial communities affect health and disease.
Hoboken, New Jersey: John Wiley & Sons, Inc; 2013. P. 211-253. doi: 10.1002/9781118409855.
33. Sung JY, Costerton JW, Shaffer EA. Defense system in the biliary tract against bacterial infection. Dig Dis Sci. 1992;37(5):689-696. doi: 10.1007/Bf01296423.
34. Robinson CM, Pfeiffer JK. Viruses and the Microbiota. Annu Rev Virol. 2014;1:55-69. doi: 10.1146/annurev-virology-031413-085550.
35. Mueller T, Beutler C, Pico AH, et al. Enhanced innate immune responsiveness and intolerance to intestinal endotoxins in human biliary epithelial cells contributes to chronic cholangitis. Liver Int. 2011;31(10):1574—1588. doi: 10.1111/j.1478-3231.2011.02635.x.
36. Ye F, Shen H, Li Z, et al. Influence of the biliary system on biliary bacteria revealed by bacterial communities of the human biliary and upper digestive tracts. PLoS One. 2016;11(3):e0150519. doi: 10.1371/journal.pone.0150519.
37. Gunn JS. Mechanisms of bacterial resistance and response to bile. Microbes Infect. 2000;2(8):907—913. doi: 10.1016/s1286-4579(00)00392-0.
38. Giannelli V, Di Gregorio V, Iebba V, et al. Microbiota and the gut-liver axis: bacterial translocation, inflammation and infection in cirrhosis. World J Gastroenterol. 2014;20(45):16795—16810. doi: 10.3748/wjg.v20.i45.16795.
39. Miyake Y, Yamamoto K. Role of gut microbiota in liver diseases. Hepatol Res. 2013;43(2):139—146. doi: 10.1111/j.1872-034X.2012.01088.x.
40. Govorun VM, Momynaliev KT, Chelisheva VV, Isakov VA. Helicobacter spp. found in gallbladder stones. Gut. 2002;51(Suppl 2):A74.
41. Gevers D, Kugathasan S, Denson LA, et al. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe. 2014;15(3):382—392. doi: 10.1016/j.chom.2014.02.005.
42. Pflughoeft KJ, Versalovic J. Human microbiome in health and disease. Annu Rev Pathol. 2012;7:99—122. doi: 10.1146/annurev-pathol-011811-132421.
43. Markland SM, LeStrange KJ, Sharma M, Kniel KE. Old friends in new places: exploring the role of extraintestinal E. coli in intestinal disease and foodborne illness. Zoonoses Public Health. 2015;62(7):491—496. doi: 10.1111/zph.12194.
44. Lozupone CA, Li M, Campbell TB, et al. Alterations in the gut microbiota associated with HIV-1 infection. Cell Host Microbe. 2013;14(3):329—339. doi: 10.1016/j.chom.2013.08.006.
45. Compare D, Coccoli P, Rocco A, et al. Gut-liver axis: The impact of gut microbiota on non alcoholic fatty liver disease. Nutr Metab Cardiovasc Dis. 2012;22(6):471-476. doi: 10.1016/j. numecd.2012.02.007.
46. Hoban A, Stilling R, Desbonnet L, et al. Regulation of myelina-tion in the prefrontal cortex by the gut microbiota: implications for health and disease. FASEB J. 2015;29(1 Suppl):672.4.
47. Backhed F, Fraser CM, Ringel Y, et al. Defining a healthy human gut microbiome: current concepts, future directions, and clinical applications. Cell Host Microbe. 2012;12(5):611-622. doi: 10.1016/j. chom.2012.10.012.
48. Varyukhina S, Freitas M, Bardin S, et al. Glycan-modifying bacteria-derived soluble factors from Bacteroides thetaiotaomi-cron and Lactobacillus casei inhibit rotavirus infection in human intestinal cells. Microbes Infect. 2012;14(3):273-278. doi: 10.1016/j. micinf.2011.10.007.
49. Kuss SK, Best GT, Etheredge CA, et al. Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science. 2011;334(6053):249-252. doi: 10.1126/science.1211057.
50. Гибадулина И.О., Гибадулин Н.В. Диагностические аспекты хронического холангита после холецистэктомии // Экспериментальная и клиническая гастроэнтерология. — 2011. — №6 — С. 68-72. [Gibadulina IO, Gibadulin NV. Diagnostic aspects of chronic cholangitis after cholecystectomy. Eksp Klin Gastroenterol. 2011;(6):68-72. (In Russ).]
51. Shaffer EA. Epidemiology and risk factors for gallstone disease: has the paradigm changed in the 21st century? Curr Gastroenterol Rep. 2005;7(2):132-140. doi: 10.1007/s11894-005-0051-8.
52. Nakanuma Y. Tutorial review for understanding of cholangiopathy. Int J Hepatol. 2012;2012:547840. doi: 10.1155/2012/547840.
53. Welch RA, Burland V, Plunkett G 3rd, et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc Natl Acad Sci U S A. 2002;99(26):17020-17024. doi: 10.1073/pnas.252529799.
54. Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207-214. doi: 10.1038/nature11234.
55. Simon C, Daniel R. Metagenomic analyses: past and future trends. Appl Environ Microbiol. 2011;77(4): 1153-1161. doi: 10.1128/ AEM.02345-10.
56. Aagaard K, Petrosino J, Keitel W, et al. The Human Microbiome Project strategy for comprehensive sampling of the human micro-biome and why it matters. FASEB J. 2013;27(3): 1012-1022. doi: 10.1096/fj.12-220806.
57. Dubinkina VB, Ischenko DS, Ulyantsev VI, et al. Assessment of k-mer spectrum applicability for metagenomic dissimilarity analysis. BMC Bioinformatics. 2016;17:38. doi: 10.1186/s12859-015-0875-7.
58. Scott AJ, Khan GA. Origin of bacteria in bileduct bile. Lancet. 1967;2(7520):790-792. doi: 10.1016/S0140-6736(67)92231-3.
59. Hov JR. The microbiome and human disease: a new organ of interest in biliary disease. In: Hirschfield G, Adams D, Liaskou E, editors. Biliary disease: from science to clinic. Cham: Springer International Publishing; 2017. Р. 85-96. doi: 10.1007/978-3-319-50168-0_5.
60. Colman RJ, Rubin DT. Fecal microbiota transplantation as therapy for inflammatory bowel disease: a systematic review and meta-analysis. J Crohns Colitis. 2014;8(12): 1569-1581. doi: 10.1016/j. crohns.2014.08.006.
61. Bolan S, Seshadri B, Talley NJ, Naidu R Bio-banking gut micro-biome samples. EMBO Rep. 2016;17(7):929-930. doi: 10.15252/ embr.201642572.
62. Tabibian JH, O'Hara SP, Lindor KD. Primary sclerosing cholangitis and the microbiota: current knowledge and perspectives on etiopathogenesis and emerging therapies. Scand J Gastroenterol. 2014;49(8):901-908. doi: 10.3109/00365521.2014.913189.
63. Mattner J. Impact of microbes on the pathogenesis of primary biliary cirrhosis (PBC) and primary sclerosing cholangitis (PSC). Int J Mol Sci. 2016;17(11):1864. doi: 10.3390/ijms17111864.
64. Vitek L, Carey MC. New pathophysiological concepts underlying pathogenesis of pigment gallstones. Clin Res Hepatol Gastroenterol. 2012;36(2):122-129. doi: 10.1016/j.clinre.2011.08.010.
65. Stinton LM, Myers RP, Shaffer EA. Epidemiology of gallstones. Gastroenterol Clin North Am. 2010;39(2): 157-169. doi: 10.1016/j. gtc.2010.02.003.
66. Keren N, Konikoff FM, Paitan Y, et al. Interactions between the intestinal microbiota and bile acids in gallstones patients. Environ Microbiol Rep. 2015;7(6):874-880. doi: 10.1111/17582229.12319.
67. Krastev Z, Vladimirov B, Mateva L, Alexiev A. Quantitative assessment of severity of biliary tract infection. Hepatogastroenterology. 1996;43(10):792-795.
68. Darkahi B, Sandblom G, Liljeholm H, et al. Biliary microflora in patients undergoing cholecystectomy. Surg Infect (Larchmt). 2014;15(3):262-265. doi: 10.1089/sur.2012.125.
69. Cotter PD, Stanton C, Ross RP, Hill C. The impact of antibiotics on the gut microbiota as revealed by high throughput DNA sequencing. Discov Med. 2012;13(70):193—199.
70. Modi SR, Collins JJ, Relman DA. Antibiotics and the gut microbiota. J Clin Invest. 2014;124(10):4212—4218. doi: 10.1172/ Jci72333.
71. Ecological modeling from time-series inference: insight into dynamics and stability of intestinal microbiota. PLoS Comput Biol. 2013;9(12):e1003388. doi: 10.1371/journal.pcbi.1003388.
72. Глебов К.Г., Котовский А.Е., Дюжева Т.Г. Критерии выбора конструкции эндопротеза для эндоскопического стентиро-вания желчных протоков // Анналы хирургической гепатоло-гии. — 2014. — T.19. — №2 — С. 55—65. [Glebov KG, Kotov-skiy AE, Dyuzheva TG. Criteria for the choice of construction of endoprosthesis for endoscopic biliary stenting. Annaly khirurgiches-koigepatologii. 2014;19(2):55-65. (In Russ.)]
73. Swidsinski A, Schlien P, Pernthaler A, et al. Bacterial biofilm within diseased pancreatic and biliary tracts. Gut. 2005;54(3):388—395. doi: 10.1136/gut.2004.043059.
КОНТАКТНАЯ ИНФОРМАЦИЯ Клабуков Илья Дмитриевич, научный сотрудник отдела передовых клеточных технологий Института регенеративной медицины Первого Московского государственного медицинского университета им. И.М. Сеченова Минздрава России
Адрес: 119991, Москва, ул. Трубецкая, д. 8, стр. 2, тел.: +7 (495) 609-14-00, e-mail: ilya.klabukov@gmail.com, SPIN-код: 3388-8859, ORCID ID: http://orcid.org/0000-0002-2888-7999
Люндуп Алексей Валерьевич, кандидат медицинских наук, заведующий отделом передовых клеточных технологий Института регенеративной медицины Первого Московского государственного медицинского университета им. И.М. Сеченова Минздрава России
Адрес: 119991, Москва, ул. Трубецкая, д. 8, стр. 2, тел.: +7 (495) 609-14-00, e-mail: lyundup@gmail.com, SPIN-код: 4954-3004, ORCID ID: http://orcid.org/0000-0002-0102-5491
Дюжева Татьяна Геннадьевна, доктор медицинских наук, профессор, заведующая отделом регенеративной хирургии печени и поджелудочной железы Института регенеративной медицины Первого Московского государственного медицинского университета им. И.М. Сеченова Минздрава России Адрес: 119991, Москва, ул. Трубецкая, д. 8, стр. 2, тел.: +7 (495) 609-14-00, e-mail: dtg679@gmail.com, SPIN-код: 7325-2086, ORCID ID: http://orcid.org/0000-0003-0573-7573
Тяхт Александр Викторович, кандидат биологических наук, научный сотрудник лаборатории биоинформатики Федерального научно-клинического центра физико-химической медицины ФМБА России Адрес: 119435, Москва, ул. Малая Пироговская, д. 1а, тел.: +7 (499) 245-94-59, e-mail: a.tyakht@gmail.com, SPIN-код: 6183-5445, ORCID ID: http://orcid.org/0000-0002-7358-2537
179
Рекомендуемые статьи
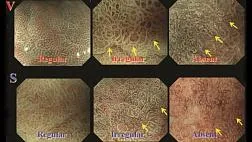
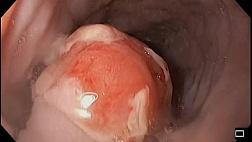
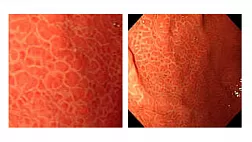
Эндоскопическая ретроградная холангиопанкреатография
- рентген-эндоскопический метод диагностики патологии желчных и панкреатических протоков.
При эндоскопическом исследовании в случае бронхоэктазов в стадии ремиссии выявляется
частично диффузный бронхит I степени воспаления
Активируйте PUSH уведомления в браузер
Отключите PUSH уведомления в браузер
Содержание
Интернет магазин
Популярное
- О нас
- Правовые вопросы
- Политика
обработки персональных
данных EndoExpert.ru - Связаться с нами
- Стать партнером
© 2016-2022 EndoExpert.ru

Вы находитесь в разделе предназначенном только для специалистов (раздел для пациентов по ссылке). Пожалуйста, внимательно прочитайте полные условия использования и подтвердите, что Вы являетесь медицинским или фармацевтическим работником или студентом медицинского образовательного учреждения и подтверждаете своё понимание и согласие с тем, что применение рецептурных препаратов, обращение за той или иной медицинской услугой, равно как и ее выполнение, использование медицинских изделий, выбор метода профилактики, диагностики, лечения, медицинской реабилитации, равно как и их применение, возможны только после предварительной консультации со специалистом. Мы используем файлы cookie, чтобы предложить Вам лучший опыт взаимодействия. Файлы cookie позволяют адаптировать веб-сайты к вашим интересам и предпочтениям.
Я прочитал и настоящим принимаю вышеизложенное, хочу продолжить ознакомление с размещенной на данном сайте информацией для специалистов.












.jpg)


.png)















Комментарии